Novel urine biomarker may inform biopsy decision

By J. Stephen Jones, MD, and Ahmad Elshafei, MD, PhD
Advertisement
Cleveland Clinic is a non-profit academic medical center. Advertising on our site helps support our mission. We do not endorse non-Cleveland Clinic products or services. Policy

J. Stephen Jones, MD

Ahmad Elshafei, MD, PhD
A new prostate cancer prediction tool developed and validated by investigators at Cleveland Clinic’s Glickman Urological & Kidney Institute achieves high accuracy, especially for high-grade tumors, by incorporating the prostate cancer antigen gene 3 (PCA3) as a biomarker.
The newly developed nomogram is biopsy protocol-specific and is based on a large cohort of men who underwent initial prostate biopsy.
Although further validation is needed, the PCA3-based nomogram has potential clinical utility to identify patients at risk for harboring prostate cancer, especially high-grade cancer, at initial biopsy.
Accurate and reliable early-detection methods for prostate cancer are needed. The prostate-specific antigen (PSA) protein, which is the most frequently used biomarker for prostate cancer, lacks specificity, often resulting in potentially unnecessary biopsy.
The PCA3 gene, formally known as the DD3 gene, is a non-coding gene located at chromosome 9q21-22 whose expression is restricted to prostate tissue. PCA3 is overexpressed in at least 94 percent of prostate tumors.
The goal of using PCA3 as a biomarker is to reduce the number of unnecessary biopsies. The literature supports the predictive accuracy of PCA3 over PSA for early detection of prostate cancer.
Prior PCA3-based predictive models were not biopsy protocol-specific as they included men who underwent initial and repeat prostate biopsy.
The goal of our retrospective study was the development and internal validation of a PCA3-based nomogram for prediction of overall prostate cancer and high-grade prostate cancer in a large North American cohort who underwent initial prostate biopsy.
Advertisement
Our cohort consisted of 3,675 patients from a single institution (Western New York Urology Associates) who had undergone transrectal ultrasound-guided prostate biopsy of at least 10 cores after detection of an elevated PSA level (≤ 20 ng/mL) and/or abnormal findings on digital rectal examination (DRE). After prostate massage, 20 to 30 mL of voided urine was collected and analyzed for PCA3 prior to biopsy.
Biopsy results confirmed prostate cancer in 44 percent of the 3,675 patients, and high-grade prostate cancer was present in 19.1 percent.
The variables whose predictive values were tested for inclusion in our nomogram were age, PSA level, PCA3 score, race, family history of prostate cancer, DRE findings and prostate volume.
We calculated subjects’ PCA3 score using the formula (PCA3 mRNA/PSA mRNA) ×1,000.
We constructed two logistic regression models to predict overall prostate and high-grade prostate cancer, defined as Gleason score ≥ 7.
The multivariate logistic regression models showed that age, PSA level, PCA3 score, prostate volume, family history of prostate cancer and abnormal DRE were independent predictors for prostate cancer overall. Age, PSA level, PCA3 score, prostate volume and abnormal DRE were independent predictors for high-grade prostate cancer.
We built a PCA3-based logistic regression nomogram using the risk factors from the logistic regression models that predicted overall prostate cancer and high-grade prostate cancer.
The PCA3-based nomogram demonstrated a predictive accuracy (concordance index) of 0.742 for prostate cancer overall and a c-index of internal validation of 0.768 when applied for the prediction of high-grade prostate cancer. The base model without PCA3 showed a decline in the c-index from 0.742 to 0.700 for the prediction of prostate cancer overall, and from 0.768 to 0.753 for the prediction of high-grade prostate cancer.
Advertisement
Agreement (calibration) of the two nomograms for prediction of overall prostate cancer and high-grade prostate cancer was excellent.
Including PCA3 in multivariate models predicting prostate cancer and high-grade prostate cancer improves the predictive accuracy of the models. PCA3 is a useful tool to identify patients at risk of prostate cancer at initial biopsy.
Dr. Jones holds the Leonard Horvitz and Samuel Miller Distinguished Chair in Urological Oncology Research.
Dr. Elshafei is a Research Associate in Cleveland Clinic’s Glickman Urological & Kidney Institute.
Advertisement
Advertisement

First-of-its-kind research investigates the viability of standard screening to reduce the burden of late-stage cancer diagnoses

Global R&D efforts expanding first-line and relapse therapy options for patients
Study demonstrates ability to reduce patients’ reliance on phlebotomies to stabilize hematocrit levels

A case study on the value of access to novel therapies through clinical trials

Findings highlight an association between obesity and an increased incidence of moderate-severe disease
Cleveland Clinic Cancer Institute takes multi-faceted approach to increasing clinical trial access 23456
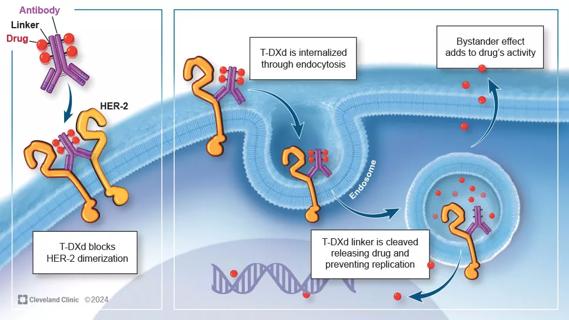
Key learnings from DESTINY trials

Overall survival in patients treated since 2008 is nearly 20% higher than in earlier patients