Data suggests “field cancerization” effect in early-stage disease
By Cristina Magi-Galluzzi, MD, PhD; Sara Falzarano, MD; and Eric A. Klein, MD
Advertisement
Cleveland Clinic is a non-profit academic medical center. Advertising on our site helps support our mission. We do not endorse non-Cleveland Clinic products or services. Policy

Eric A. Klein, MD

Cristina Magi-Galluzzi, MD, PhD

Sara Falzarano, MD
Development of biomarkers that can accurately predict the biologic potential of early-stage prostate cancer has been challenging because of the heterogeneity in grade and multifocality of most tumors.
One approach to addressing these challenges is to incorporate a deeper understanding of the biology of the nonmalignant stromal and epithelial cells present within and adjacent to the tumor, which may represent a generalized field effect related to tumor aggressiveness.
The concept of field cancerization has evolved, from an initial histological definition credited to Slaughter, et al, to a molecular definition exemplified by a range of genetic and epigenetic abnormalities that can be detected in normal-appearing tissues adjacent to cancers, including gene silencing by methylation, deletions in mitochondrial DNA, mutations in cancer-related genes, and aberrant gene expression. Evidence for a field effect has been reported in a variety of cancer types, including head and neck, lung, colon, breast, stomach, and bladder.
Advertisement
We previously reported that a validated 17-gene expression assay, the Genomic Prostate Score (GPS, owned and marketed as OncotypeDx Prostate by Genomic Health, Inc.) provides an independent biologic measure of tumor aggressiveness that improves risk assessment for men with newly diagnosed, clinically low-risk prostate cancer. This assay has been shown to influence clinical decision-making by distinguishing men with biologically nonaggressive tumors who are appropriate candidates for active surveillance from men with more aggressive tumors who should be considered for immediate therapy.
A pre-specified aim of the original gene identification study that led to development of GPS conducted on radical prostatectomy specimens was to assess gene expression in normal tissue adjacent to prostate tumor, where “adjacent” was defined as ≥ 3 mm distant from tumor.
The goal of the present analysis was to identify gene expression changes in normal tissue that could predict clinical outcome and to compare the gene expression patterns from normal to tumor tissue. Additionally, post-hoc analyses were conducted to evaluate if GPS, developed using the tumor-containing tissue, was predictive of outcome when applied to gene expression derived from normal specimens.
In this latest study we asked whether identification of genes in normal tissue would provide evidence for a biologically meaningful field effect in the tumor-containing prostate and identify early markers of aggressive prostate cancer assayable in non-tumor tissue.
Advertisement
For this study, 501 prostatectomy specimens were centrally reviewed by two urologic pathologists to assess primary and secondary/tertiary Gleason patterns and overall Gleason score using the 2005 International Society of Urological Pathology Consensus guidelines.
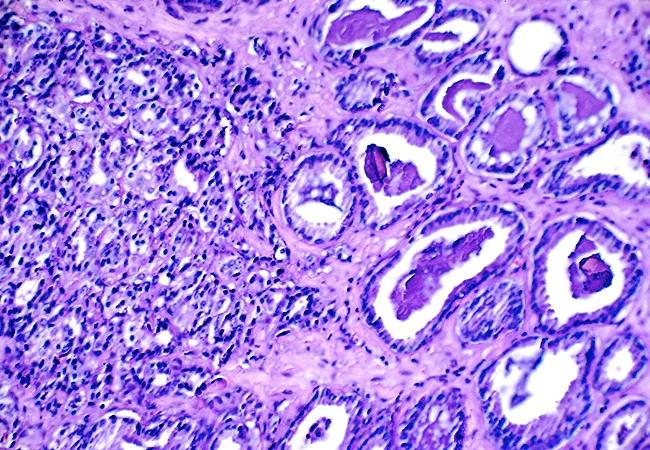
For each patient, two spatially distinct tumor specimens, representing the primary Gleason pattern and the highest Gleason pattern, were manually microdissected using a microscope. A single area of normal tissue adjacent to but at least ≥ 3 mm from the cancer was also microdissected (see Figure 1). Gene expression was measured by RT-PCR; contamination of normal tissue by tumor was excluded by the absence of TRMPSS2-ERG fusions.

Figure 1. Mapping and microdissection of tumor and normal tissue. From Magi-Galluzzi C, Maddala T, Falzarano SM, et al. Gene expression in normal-appearing tissue adjacent to prostate cancers are predictive of clinical outcome: evidence for a biologically meaningful field effect. Oncotarget. 2016 Apr 22 [Epub ahead of print].
Of 732 candidate cancer-related genes assayed, five genes were excluded due to poor analytical performance, yielding 727 evaluable genes. Controlling the false discovery rate at 20 percent, 46 genes measured in normal tissue predicted clinical recurrences (local or distant failure or prostate cancer-related death), indicating that gene expression patterns in the normal tissue adjacent to prostate cancer can predict meaningful clinical outcomes.
Among these 46 genes, 39 (85 percent) were associated with clinical recurrence in tumor tissues as well. The genes associated with clinical recurrence in the normal tissue represent a diverse range of biological pathways (see Table).
Advertisement

Table. Functional groups of the 39 genes associated with clinical recurrence in normal tissue
Higher expression of stromal response and proliferation genes was associated with higher risk of clinical recurrence, whereas, for the androgen signaling, cellular organization, chromatin remodeling, protein folding and stress response gene groups, reduced expression was associated with higher recurrence risk.
The standardized hazard ratios for these genes ranged from 0.6 to 0.8 for genes associated with better outcome and 1.3 to 1.6 for genes associated with worse outcome; for a majority of these genes (77 percent; 30/39), the association was such that down-regulated gene expression was associated with worse outcome. For 33 of these 39 genes, the association of gene expression with clinical recurrence was directionally consistent in normal tissue and tumor, although a stronger association was usually observed in tumor compared to normal tissue (see Figure 2).

Figure 2. Forest plot comparing of strength of genes in predicting clinical recurrence when assessed in adjacent normal-appearing (NT) and tumor tissues (primary or highest Gleason pattern). Standardized hazard ratios are shown as red triangles for the primary Gleason pattern tumor, orange circles for the highest Gleason pattern tumor, and blue squares for the adjacent NT.
The results support the existence of a biologically meaningful field cancerization in prostate cancer, demonstrating that gene expression patterns in normal tissue adjacent to cancer can predict clinical outcome, and that assessing gene expression in normal tissue may help identify those patients with aggressive disease.
Advertisement
Furthermore, the tumor-based 17-gene GPS, which was derived from tumor samples within this dataset, was associated with clinical outcome when measured in normal tissue, although the strength of association was weaker than in tumor.
The results should advance the understanding of prostate cancer biology and aid cancer detection. They also have implications for the success or failure of therapies using subtotal gland ablation, which generally leave normal-appearing tissue that may have biological potential untreated.
This research was performed in collaboration with Genomic Health, Inc., which provided funding for the study.
Advertisement

First-of-its-kind research investigates the viability of standard screening to reduce the burden of late-stage cancer diagnoses

Global R&D efforts expanding first-line and relapse therapy options for patients
Study demonstrates ability to reduce patients’ reliance on phlebotomies to stabilize hematocrit levels

A case study on the value of access to novel therapies through clinical trials

Findings highlight an association between obesity and an increased incidence of moderate-severe disease
Cleveland Clinic Cancer Institute takes multi-faceted approach to increasing clinical trial access 23456
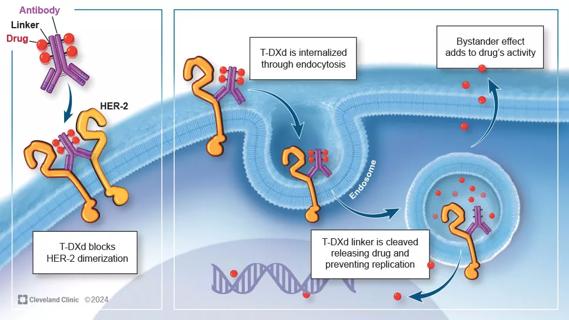
Key learnings from DESTINY trials

Overall survival in patients treated since 2008 is nearly 20% higher than in earlier patients